|
|
Diffusion Accessibility
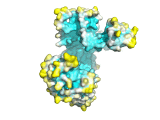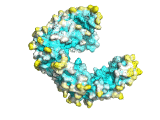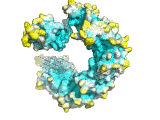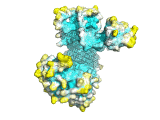
Diffusion accessibility gives a measure of surface accessibility that is longer range in nature than ordinary solvent accessibility. The idea of diffusion accessibility was introduced several years ago (Yeates, TO (1995) Algorithms for Evaluating the Long-range Accessibility of Protein Surfaces. J Mol Biol 249(4): 804-15). Diffusion accessibility is a measure of how easily or frequently a specific point on a surface would capture a hypothetical diffusing (or randomly-walking) probe that is captured upon its first encounter with any part of a surface. Naturally, parts of a surface that are solvent-exposed in the traditional sense but lie in a deep surface depression will capture the probe only rarely, since the probe will more often encounter another part of the surface first. The results of a diffusion accessibility calculation can be useful in quantitatively evaluating potential binding regions of a protein surface, and are particularly useful in visualization. Even the most widely used graphics programs for visualizing proteins and their surfaces generally do not produce images that make the depth and character of protein surfaces very obvious. Using the diffusion accessibility to color a surface can add substantially to the depth cues and can produce a better appreciation of a surface, particularly those regions likely to be involved in binding.
|
As discussed in the 1995 paper, the best way to calculate diffusion accessibility is to cast it as a problem in steady-state diffusion, and solve the resulting (Poisson) equations on a 3D grid. This website performs that calculation on an uploaded PDB file. The calculation is performed on the server side and can be accessed here. The program returns another pdb file that contains a normalized value of the diffusion accessibility for each atom in the temperature factor (B factor) column. The best way to use that information to color your images is to use PyMol to view the new PDB file, and then color it using a PyMol script specially written for that purpose.
Instructions for use:
- Click on "Do Calculation". There you will upload a PDB file and retrieve a modified version with the required information in the temperature factor column.
- Click on "Get Coloring Script" to download the PyMol script. You need only do this once, not each time you analyze a new PDB file. Information about running the script is located at that link.
An example of the program applied to the MHC peptide binding domain is shown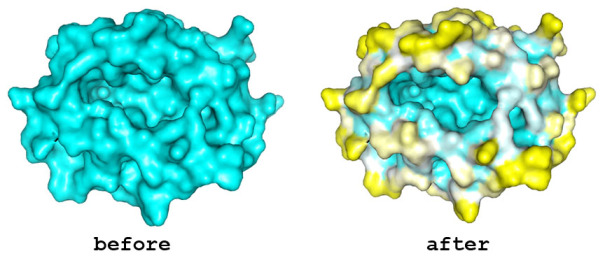
In order to run the program locally (e.g. to apply it to many pdb files) the Fortran program can be downloaded here.
Yingssu Tsai was responsible for developing the PyMol script. Tom Holton maintains the server. The diffusion accessibility calculation was written by Todd Yeates.